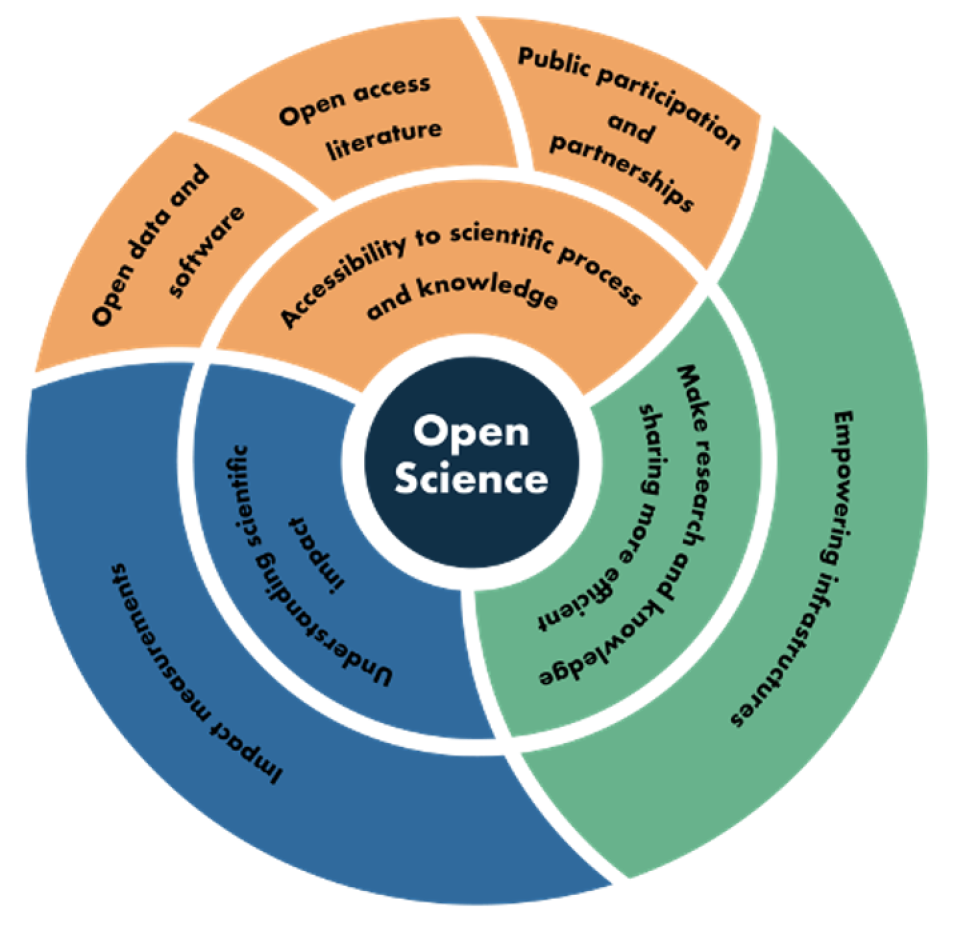
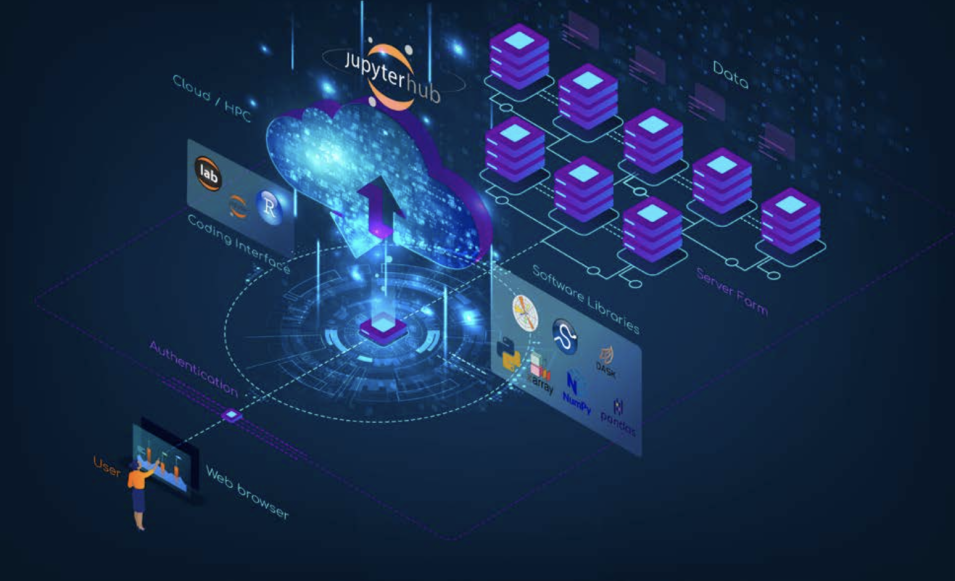
Transformational. This is how Dr. Chelle Gentemann describes her work with openly available NASA Earth observing data using open source software. Gentemann, a physical oceanographer and the open science lead for NASA’s Earth Science Data Systems (ESDS) Program, has seen her use of data – and what she can do with these data – evolve significantly over the past few years. Her journey adapting to this new paradigm of collaborative, openly sourced science using cloud-based Big Data collections is one being undertaken by scientists and researchers around the globe. This is leading to a shift in not only how science is conducted, but the skills scientists need to succeed in this new environment. As stewards of NASA Earth observing data, ESDS is committed to guiding users in this paradigm shift to enable the most efficient use of data in NASA’s Earth Observing System Data and Information System (EOSDIS) collection.
In recently-published peer-reviewed article in AGU Advances (doi:10.1029/2020AV000354), Gentemann and her co-authors discuss this evolution in the core tools of science – data, software, and computers – and how this is enabling collaborative, interdisciplinary science wherever and whenever an internet connection is available. As she observes, this paradigm shift in how science is conducted and in how data are used is a two-way street, requiring evolution by both data users and data providers.
Let’s start with your journey as a scientist. What was your path to using open source software in your research?
For many years, I did research funded with federal grants for a private research company. When I left they claimed ownership of all my software. At that point, I was fairly well along in my career, and I had a decision to make. Do I keep doing science the same way and re-write my old software or do I try something new? I had been hearing all of this stuff about Python and open source software, so I took a risk and decided to try Python. Initially, I was writing all of my code from scratch, just like I used to in Fortran.
As I started to learn about the open source ecosystem, it all suddenly clicked. You have that moment where you’re like, oh my gosh, there’s a reason why open source software is the language of science for the future. It’s because instead of having to write a new program for every type of netCDF file, I could use the software library Xarray and it could read any netCDF file in one line. Additionally, I could read all the files in a dataset in one line because it was a more advanced software tool. I could read not only netCDF, but many different types of files easily because open source developers had already written generalized software. I was just shell-shocked for a while and so incredibly amazed at what I hadn’t known was available.
What are some of the benefits you’ve experienced from your use of open source software?
[Open source software] accelerates my science. Two weeks ago, a colleague was trying to recreate a figure that I had published in a paper about 10 years ago. A document was due the next day and they needed to know a cutoff point in the data. And I said, oh, don’t use that data, it’s 10 years old. This was not a small dataset; it was a global, 30-year, 25-kilometer dataset produced four times a day. [Producing] the original figure took me probably three weeks and hundreds of lines of code, not even counting the time to download the data – which would add two or three weeks. Using [open source software], it took me about 30 minutes to recreate the figure and give him the exact cutoff point with the new data.
Being able to switch from something that took about a month to being able to do the same work in 30 minutes allows me to do more data investigation, it allows me to explore new ideas, and it allows me to be much more agile in how I do my science. And that’s why I like open source software. It is transformational and it’s more collaborative.