Longtime readers of this blog know IMPACT’s research team explores diverse topics in Earth science. Recently, however, researchers from the machine learning team have expanded their field of interest to collaborate with NASA’s Biological and Physical Sciences (BPS) division. In order to better understand the physiological changes that occur as a consequence of spaceflight, mice are sent to the International Space Station and housed for variable periods of time before their return to Earth.
As part of this effort, NASA GeneLab is an open-source data repository for “omics” data (omics is a multi-disciplinary field encompassing genomics, epigenomics, transcriptomics, proteomics, and metabolomics) from spaceflight and space-relevant biological experiments, while NASA Ames Life Sciences Data Archive (ALSDA) curates and shares the non-omic and environmental data from experiments.
In order to facilitate future algorithm development for space biology, and to encourage standardized benchmarking of methods for specific problems, IMPACT researchers Nishan (Nish) Pantha and Vishal Perekadan leveraged two common types of biological data from GeneLab and ALSDA: gene expression (synthetic RNA transcript data) and microscopy (space radiation immune cell microscopy benchmark dataset). Nish and Vishal presented their research last December at the American Geophysical Union 2022 Fall Meeting in Chicago.
Nish’s work leveraged machine learning (ML) algorithms to identify important genes contributing to the physical characteristics of rodents, while Vishal’s work compared convolutional neural network (CNN) and support vector machine (SVM) approaches for space radiation classification.
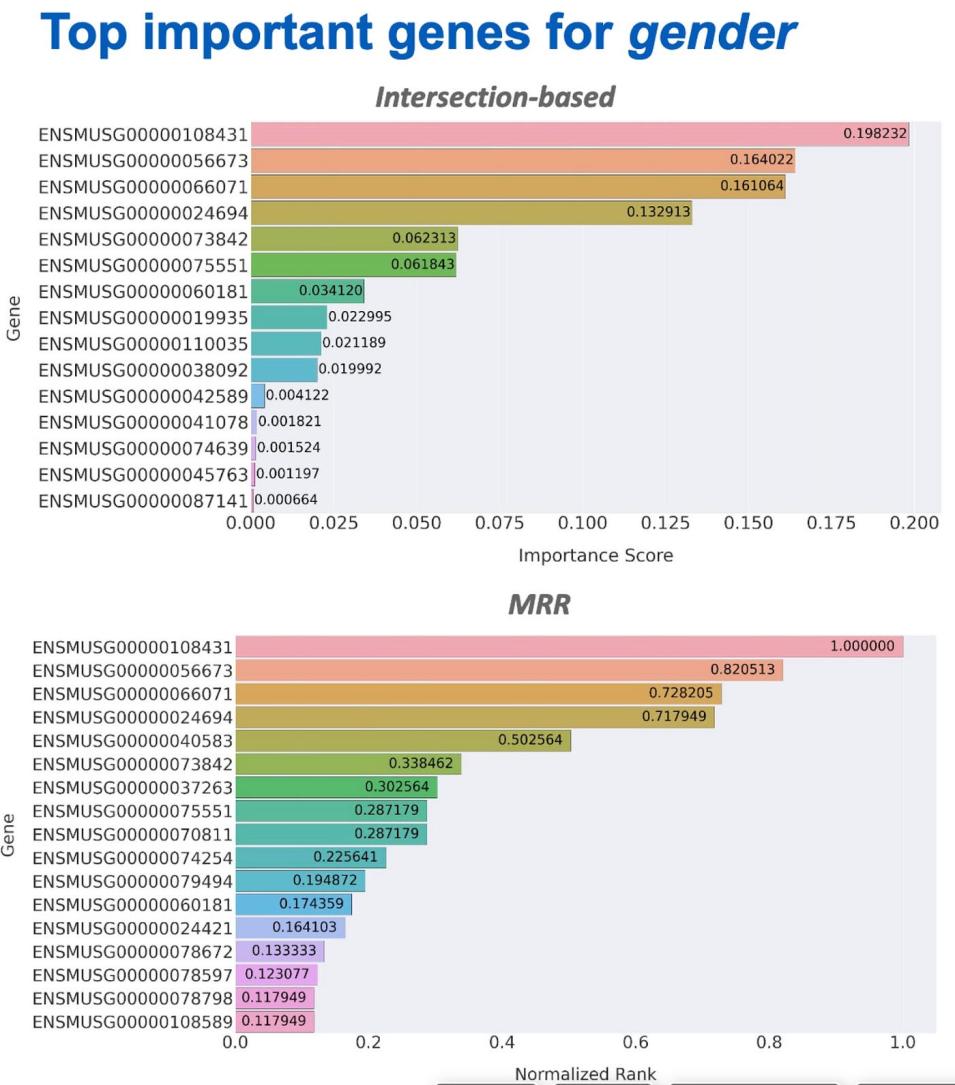
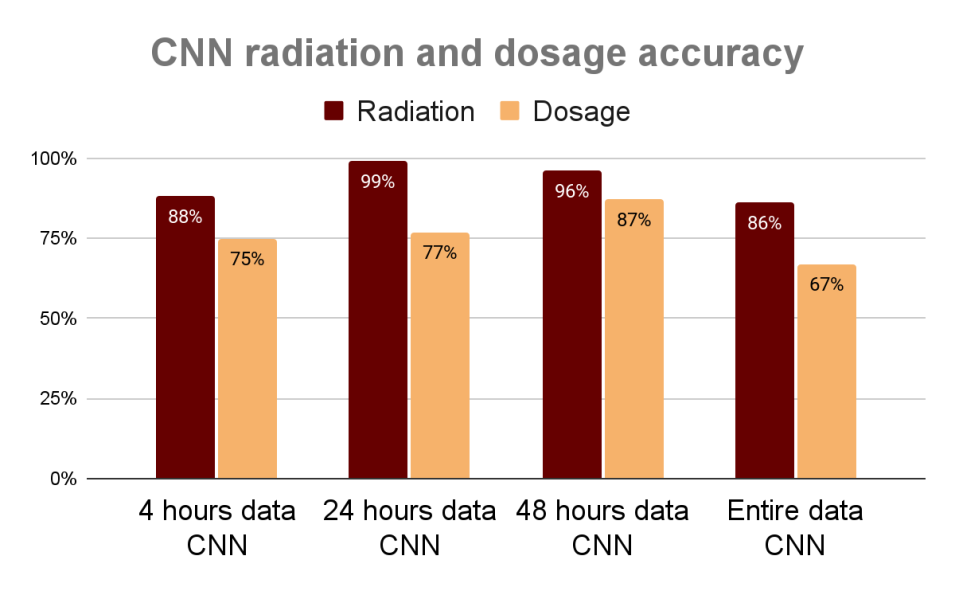
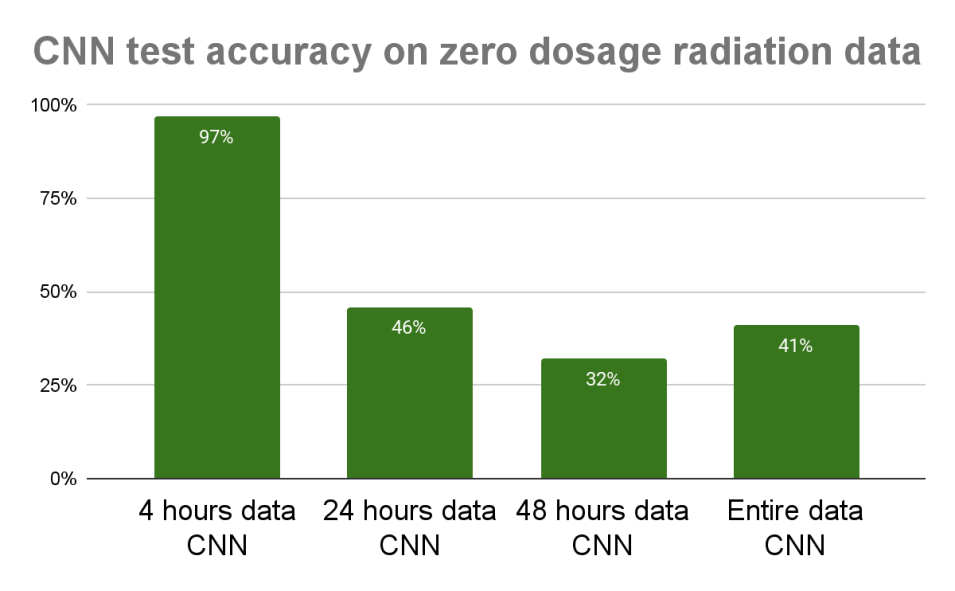
Nish and his IMPACT teammates developed an ensemble-based gene-ranking pipeline called GeneRanker that used eXtreme Gradient Boosting (XGboost) based models to identify the most important genes contributing to a target attribute. Vishal’s team’s research used a dataset of ML-ready microscopic imagery of the radiated cells, including labels indicating radiation type and dosage of exposure. These efforts focused on creating multiple benchmark models, starting with simple thresholding techniques then advancing to deep learning for radiation and dosage labeled classification.
Nish predicts his research will help the BPS team establish an initial benchmark for gene expression and could encourage the community to explore other paradigms pertaining to the feature ranking problem. Moreover, bioinformatics scientists will be able to leverage the ranking pipeline and clustering algorithms. These IMPACT teams have developed interpretable feature selection methods and ranking algorithms, particularly using ensembles of tree-based models.