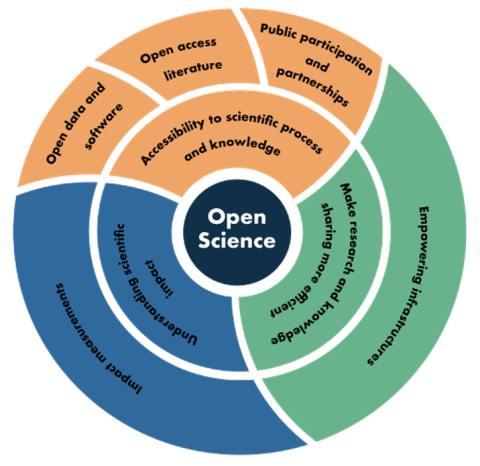
Transformational. This is how Dr. Chelle Gentemann describes her work with openly available NASA Earth observing data using open source software. Gentemann, a physical oceanographer and the open science lead for NASA’s Earth Science Data Systems (ESDS) Program, has seen her use of data – and what she can do with these data – evolve significantly over the past few years. Her journey adapting to this new paradigm of collaborative, openly sourced science using cloud-based Big Data collections is one being undertaken by scientists and researchers around the globe. This is leading to a shift in not only how science is conducted, but the skills scientists need to succeed in this new environment. As stewards of NASA Earth observing data, ESDS is committed to guiding users in this paradigm shift to enable the most efficient use of data in NASA’s Earth Observing System Data and Information System (EOSDIS) collection.
In recently-published peer-reviewed article in AGU Advances (doi:10.1029/2020AV000354), Gentemann and her co-authors discuss this evolution in the core tools of science – data, software, and computers – and how this is enabling collaborative, interdisciplinary science wherever and whenever an internet connection is available. As she observes, this paradigm shift in how science is conducted and in how data are used is a two-way street, requiring evolution by both data users and data providers.
Let’s start with your journey as a scientist. What was your path to using open source software in your research?
For many years, I did research funded with federal grants for a private research company. When I left they claimed ownership of all my software. At that point, I was fairly well along in my career, and I had a decision to make. Do I keep doing science the same way and re-write my old software or do I try something new? I had been hearing all of this stuff about Python and open source software, so I took a risk and decided to try Python. Initially, I was writing all of my code from scratch, just like I used to in Fortran.
As I started to learn about the open source ecosystem, it all suddenly clicked. You have that moment where you’re like, oh my gosh, there’s a reason why open source software is the language of science for the future. It’s because instead of having to write a new program for every type of netCDF file, I could use the software library Xarray and it could read any netCDF file in one line. Additionally, I could read all the files in a dataset in one line because it was a more advanced software tool. I could read not only netCDF, but many different types of files easily because open source developers had already written generalized software. I was just shell-shocked for a while and so incredibly amazed at what I hadn’t known was available.
What are some of the benefits you’ve experienced from your use of open source software?
[Open source software] accelerates my science. Two weeks ago, a colleague was trying to recreate a figure that I had published in a paper about 10 years ago. A document was due the next day and they needed to know a cutoff point in the data. And I said, oh, don’t use that data, it’s 10 years old. This was not a small dataset; it was a global, 30-year, 25-kilometer dataset produced four times a day. [Producing] the original figure took me probably three weeks and hundreds of lines of code, not even counting the time to download the data – which would add two or three weeks. Using [open source software], it took me about 30 minutes to recreate the figure and give him the exact cutoff point with the new data.
Being able to switch from something that took about a month to being able to do the same work in 30 minutes allows me to do more data investigation, it allows me to explore new ideas, and it allows me to be much more agile in how I do my science. And that’s why I like open source software. It is transformational and it’s more collaborative.
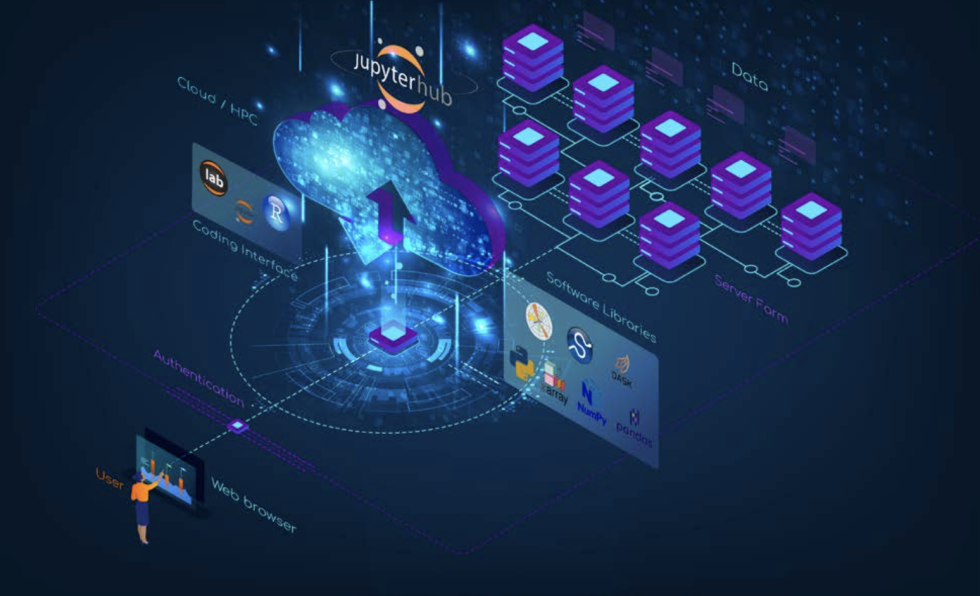
The nice thing about this whole open science ecosystem is that it’s not just the software. It’s powered by the open data that NASA provides. And as NASA is moving all of these open data onto the cloud, the applications these data can be included in are going to explode. And then you’ll have the data right next to where you can do your compute.
Traditionally, I’ve worked at very small private and nonprofit companies. And it was always sort of hard, because other groups at institutions would have these vast data resources and computational infrastructure that I never had. Now that advantage is reduced because everyone can access the cloud. Yes, you have to pay for it, but it’s inexpensive compared to setting up and maintaining a $60,000 computer at your institution. Someone can log on anywhere in the world and upload a program; they don’t need to download the data, they don’t need to buy big computers, they don’t need to have the tech infrastructure.
This also broadens who can participate in science, because there’s still a lot of countries and even parts of the United States that don’t have good internet infrastructure. And downloading data and getting computers is hard, and it’s expensive, and it’s slow. If you can bring your compute to the data, it doesn’t matter where in the world you are; you can code on a cell phone. It opens up the participation of science to anyone.
How does traditional academic training need to adapt to best prepare the next generation of scientists for working with Big Data in this open science environment?
When I was an undergraduate, research ethics, open source science, none of that was really covered, as some of it is today. I had one programming class and that was it. I think it’s really important to try to educate our incoming workforce or incoming scientists on how to do open science, because it’s still not super easy or clear. What are the ethics behind open science between who shares what and what you’re allowed to use? If you use someone’s code, are you collaborating with them? When is it okay just to give them credit? I think that these are conversations that are important to have between mentors and mentees and to have within graduate schools and undergraduate schools; we need to talk about the ethics of using open science resources, and how to do that and participate in the open science community.
I think that training scientists on some of the basics of doing open science, like how to use GitHub, how to publish in open access journals, how to create a containerized version of code, are all skills that that are going to help advance science. It will help advance their careers if [students] know how to participate in this community.
How can data users and scientists best manage this increase in the volume, velocity, and variety of data to avoid information overload?
It used to be that data were hard to get. You had to have some basic understanding of how to read different data formats. And that limited who could use them.
Open source tools are [simplifying] both the structure and access to the data so that I can almost just say “get ocean temperature,” and [the computer] is going to bring back ocean temperature data. The benefit of this is it makes it really easy to get lots of diverse datasets. The other side of that coin is that you have people using data who may not be familiar with a particular dataset’s weaknesses and strengths. They may use [the data] in a way where they’re not able to interpret their results correctly because they aren’t familiar with some of the artifacts that were in the data. When getting data is so easy, you often just assume that the data are correct.
The transmission of information about data uncertainty and about a dataset’s strengths and weaknesses is something that I think the [NASA EOSDIS] DAACs [Distributed Active Archive Centers] are really going to have to prioritize. They already do this for their datasets, but as access becomes easier, the DAACs are going have to find new ways to communicate this information to users. And of course, it’s all in the metadata. But again, you have to know to look for it. So how do we promote the interdisciplinary use of these datasets and at the same time communicate what best practices are with datasets? The DAACs are going play a really important role in doing this.
To follow up on this, what do scientists need from data providers, like the EOSDIS DAACs, to work within this evolving paradigm and make it easier to work with openly sourced data?
The datasets that scientists can work with most easily are called ARCO – that’s Analysis-Ready data, Cloud Optimized. ARCO datasets are transforming how we think of a dataset. The way we store data in the cloud is going to influence what scientists can do with them.
It also comes down to decisions, even like what type of internal chunking you do. Chunking is how you internally store the dataset to enable faster access. Do you do chunking in long time series? Do you do equal time and space chunking? Do you do spatial chunking? That influences what you’re able to do with your science and how you’re able to scale it. It’s the same data, it’s just stored a different way.
We are just beginning this next step of optimizing data for science. In the past, we’d have our golden copy [of a dataset], and that’s what everybody used. It’s going be a shift in how we think about what it means for a data archive to have a golden copy, but then also have these ARCO datasets and maybe several different versions of them to allow for different types of science. And it’s going to be very important to have those [datasets] come from trusted sources with data provenance information attached.
In your article, you and your co-authors note that “as scientific analyses are moved to the cloud, it is important that we do not recreate the same barriers that researchers currently experience with local computer clusters.” What are some of these barriers? And how can we avoid them?
Communication can be a barrier. Software developers speak a language, oceanographers speak a language, meteorologists speak a language; trying to speak in terms that anyone in any field can understand is one of the hardest things about communicating. And that’s what we really need to work on. Because if we continue to speak in our domain-specific languages, it excludes people from outside that domain to access our information.
Right now, access to the cloud is evolving rapidly. There are a number of companies working to easily provide this type of access. Some institutions have already set up institutional JupyterHubs. Scientists at these institutions now have an advantage because other people just don’t know how to get onto the cloud. Setting up a JupyterHub and environment to work, managing access and security, is fairly technical, and that’s why these companies are sort of springing up.
We have to make sure that access to science isn’t restricted to just those who can pay for it and have some pathway for other people to have access to cloud computing. One possible way to do this is binder links, where there’s an ephemeral access to cloud resources. We need to make sure that those continue to be supported so that we can provide this access to anyone and it doesn’t become a barrier.
Along with open data, you’ve noted the advantages of open source software libraries. What are some of the benefits of using software based on an open source language?
Everyone has a different science workflow; the advantage of working with open source tools is that people start contributing code to those tools. So that instead of having everybody figure out how to do a weighted mean, now there’s a weighted mean function, or there’s a group-by function that will group data by months, or seasons, or years, or however you want to group it by. Scientists are contributing their workflows and their ideas to what’s useful.
I don’t have to derive and write a calculator for my Weibull coefficients, I can go and get that from [an open source] library; I don’t have to write the code to do linear regression that’s already written in SciPy. If I want to smooth the data, that’s written in another open source library. And that makes it really easy to do science; everything just turns into calls to these libraries. One result is that your code is much simpler. It’s easier to read, it’s easier to understand, it’s less buggy because there are fewer lines of code. And so your results are more accurate and they’re faster and they’re easier.
Where do you see cloud-based open science heading? How do you see yourself conducting science 10 to 20 years from now?
My hope is that the next generation of scientists is going to see data in a completely different way. I think they’re going be asking completely different questions, and I think science will be more interdisciplinary, more application oriented, and much faster than we’ve ever seen before. I think there will be a transformation in how we think about data. And by abstracting all of that structure and access, we can allow scientists to really focus on being scientists.
Are you looking forward to this evolving paradigm?
Oh, yes! I think that the more diverse voices that we have finding solutions, the better solutions we are going to find.